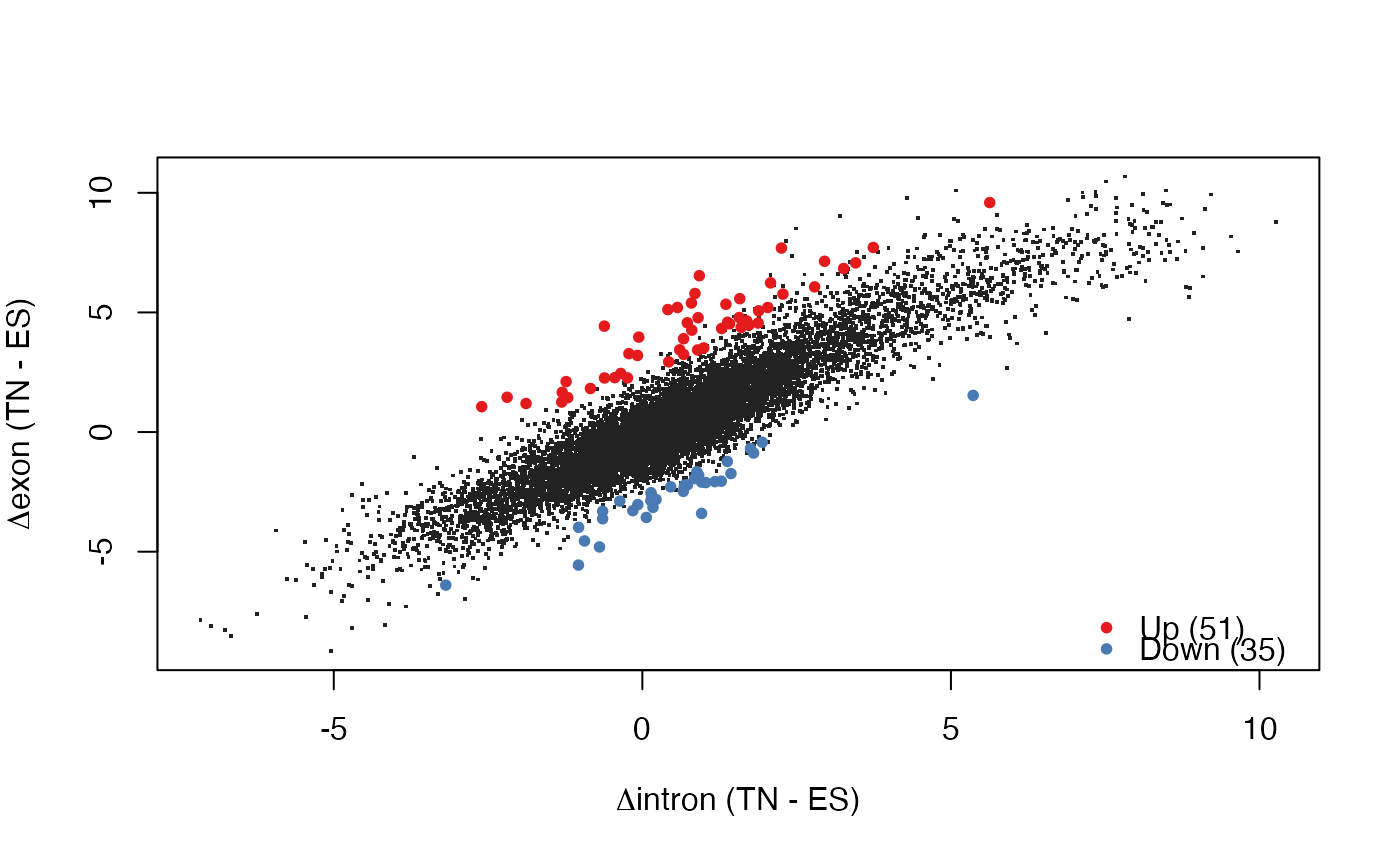
Starting from count tables with exonic and intronic counts for two conditions, perform all the steps in EISA (normalize, identify quantifyable genes, calculate contrasts and their significance).
runEISA(
cntEx,
cntIn,
cond,
method = NULL,
modelSamples = TRUE,
geneSelection = c("filterByExpr", "none", "Gaidatzis2015"),
statFramework = c("QLF", "LRT"),
legacyQLF = FALSE,
effects = c("predFC", "Gaidatzis2015"),
pscnt = 2,
sizeFactor = c("exon", "intron", "individual"),
recalcNormFactAfterFilt = TRUE,
recalcLibSizeAfterFilt = FALSE,
...
)Arguments
- cntEx
Gene by sample
matrixwith exonic counts, OR aSummarizedExperimentwith two assays namedexonandintron, containing exonic and intronic counts, respectively. IfcntExis aSummarizedExperiment,cntInwill be disregarded.- cntIn
Gene by sample
matrixwith intronic counts. Must have the same structure ascntEx(same number and order of rows and columns) ifcntExis a matrix. Will be disregarded ifcntExis aSummarizedExperiment.- cond
numeric,characterorfactorwith two levels that groups the samples (columns ofcntExandcntIn) into two conditions. The contrast will be defined as secondLevel - firstLevel.- method
One of
NULL(the default) or"Gaidatzis2015". If"Gaidatzis2015", gene filtering, statistical analysis and calculation of contrasts is performed as described in Gaidatzis et al. 2015, and the statistical analysis is based onglmFitandglmLRT. This is done by setting the argumentsmodelSamples,geneSelection,effects,pscnt,statFramework,sizeFactor,recalcNormFactAfterFiltandrecalcLibSizeAfterFiltto appropriate values (see details), overriding the defaults or any value passed to these arguments. IfNULL, the default values of the arguments will be used instead (recommended).- modelSamples
Whether to include a sample identifier in the design matrix of the statistical model. If
TRUE, potential sample effects that affect both exonic and intronic counts of that sample will be taken into account, which could result in higher sensitivity (default:TRUE).- geneSelection
Controls how to select quantifyable genes. One of the following:
"filterByExpr":(default) First, counts are normalized using
calcNormFactors, treating intronic and exonic counts as individual samples. Then,filterByExpris used with default parameters to select quantifyable genes."none":This will use all the genes provided in the count tables, assuming that an appropriate selection of quantifyable genes has already been done.
"Gaidatzis2015":First, intronic and exonic counts are linearly scaled to the mean library size (estimated as the sum of all intronic or exonic counts, respectively). Then, quantifyable genes are selected as the genes with counts
xthat fulfilllog2(x + 8) > 5in both exons and introns.
- statFramework
Selects the framework within
edgeRthat is used for the statistical analysis. One of:"QLF":(default) Quasi-likelihood F-test using
glmQLFitandglmQLFTest. This framework is highly recommended as it gives stricter error rate control by accounting for the uncertainty in dispersion estimation."LRT":- .
- legacyQLF
Whether to use the 'legacy' version of
glmQLFit. SeeglmQLFitfor more details. IfFALSE, the new method introduced inedgeR4.0.0 is used.- effects
How the effects (contrasts or log2 fold-changes) are calculated. One of:
"predFC":(default) Fold-changes are calculated using the fitted model with
predFCwithprior.count = pscnt. Please note that if a sample factor is included in the model (modelSamples=TRUE), effects cannot be obtained from that model. In that case, effects are obtained from a simpler model without sample effects."Gaidatzis2015":Fold-changes are calculated using the formula
log2((x + pscnt)/(y + pscnt)). Ifpscntis not set to 8,runEISAwill warn that this deviates from the method used in Gaidatzis et al., 2015.
- pscnt
numeric(1)with pseudocount to add to read counts (default: 2). Formethod = "Gaidatzis2015", it is set to 8. It is added to scaled read counts used ingeneSelection = "Gaidatzis2015"andeffects = "Gaidatzis2015", or else used incpm(..., prior.count = pscnt)andpredFC(..., prior.count = pscnt).- sizeFactor
How the size factors are calculated in the analysis. If 'exon' (default), the exon-derived size factors are used also for the columns corresponding to intronic counts. If 'intron', the intron-derived size factors are used also for the columns corresponding to exonic counts. If 'individual', column-wise size factors are calculated.
- recalcNormFactAfterFilt
Logical, indicating whether normalization factors should be recalculated after filtering out lowly expressed genes.
- recalcLibSizeAfterFilt
Logical, indicating whether library sizes should be recalculated after filtering out lowly expressed genes.
- ...
additional arguments passed to the
DGEListconstructor, such aslib.sizeorgenes.
Value
a list with elements
- fracIn
fraction intronic counts in each sample
- contrastName
contrast name
- contrasts
contrast matrix for quantifyable genes, with average log2 fold-changes in exons (
Dex), in introns (Din), and average difference between log2 fold-changes in exons and introns (Dex.Din)- DGEList
DGEListobject used in model fitting- tab.ExIn
statisical results for differential changes between exonic and intronic contrast, an indication for post-transcriptional regulation.
- contr.ExIn
contrast vector used for testing the difference between exonic and intronic contrast (results in
tab.ExIn)- designMatrix
design matrix used for testing the difference between exonic and intronic contrast (results in
tab.ExIn)- params
a
listwith parameter values used to run EISA
Details
Setting method = "Gaidatzis2015" has precedence over other
argument values and corresponds to setting:
modelSamples = FALSE, geneSelection = "Gaidatzis2015",
statFramework = "LRT", effects = "Gaidatzis2015", pscnt = 8,
sizeFactor = "individual", recalcNormFactAfterFilt = TRUE,
recalcLibSizeAfterFilt = FALSE.
References
Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Dimos Gaidatzis, Lukas Burger, Maria Florescu and Michael B. Stadler Nature Biotechnology, 2015 Jul;33(7):722-9. doi: 10.1038/nbt.3269.
See also
DGEList for DGEList object construction,
calcNormFactors for normalization,
filterByExpr for gene selection,
glmFit and glmQLFit for statistical
analysis.
Examples
cntEx <- readRDS(system.file("extdata", "Fig3abc_GSE33252_rawcounts_exonic.rds",
package = "eisaR"))[,-1]
cntIn <- readRDS(system.file("extdata", "Fig3abc_GSE33252_rawcounts_intronic.rds",
package = "eisaR"))[,-1]
cond <- factor(c("ES","ES","TN","TN"))
res <- runEISA(cntEx, cntIn, cond)
#> filtering quantifyable genes...
#> keeping 11759 from 20288 (58%)
#> fitting statistical model...
#> done
#> calculating log-fold changes...
#> done
plotEISA(res)
#> identified 139 genes to highlight