Query the contrast database with a set of contrasts
queryWithContrasts.RdQuery the contrast database with a set of contrasts
Arguments
- contrasts
A
SummarizedExperimentobject with assays containing contrasts named INPUT_CONTRASTS, DECODED_CONTRASTS and RESIDUAL_CONTRASTS (at least one should be present) and context information in an assay named CONTEXT. The latter is only required when use="expressed.in.both". This is typically generated usingdecomposeVar.- use
Determines if all.genes or genes expressed in both query and target context will be used. Note that "expressed.in.both", though more accurate, is slower.
- exprThr
is the quantile in the provided context that determines the expression value above which a gene is considered to be expressed. This same value is then used for thresholding the contrast database. Only applies when use="expressed.in.both".
- organism
Uses the `orthosData` contrast database from this species. One of
"Human"or"Mouse".- plotType
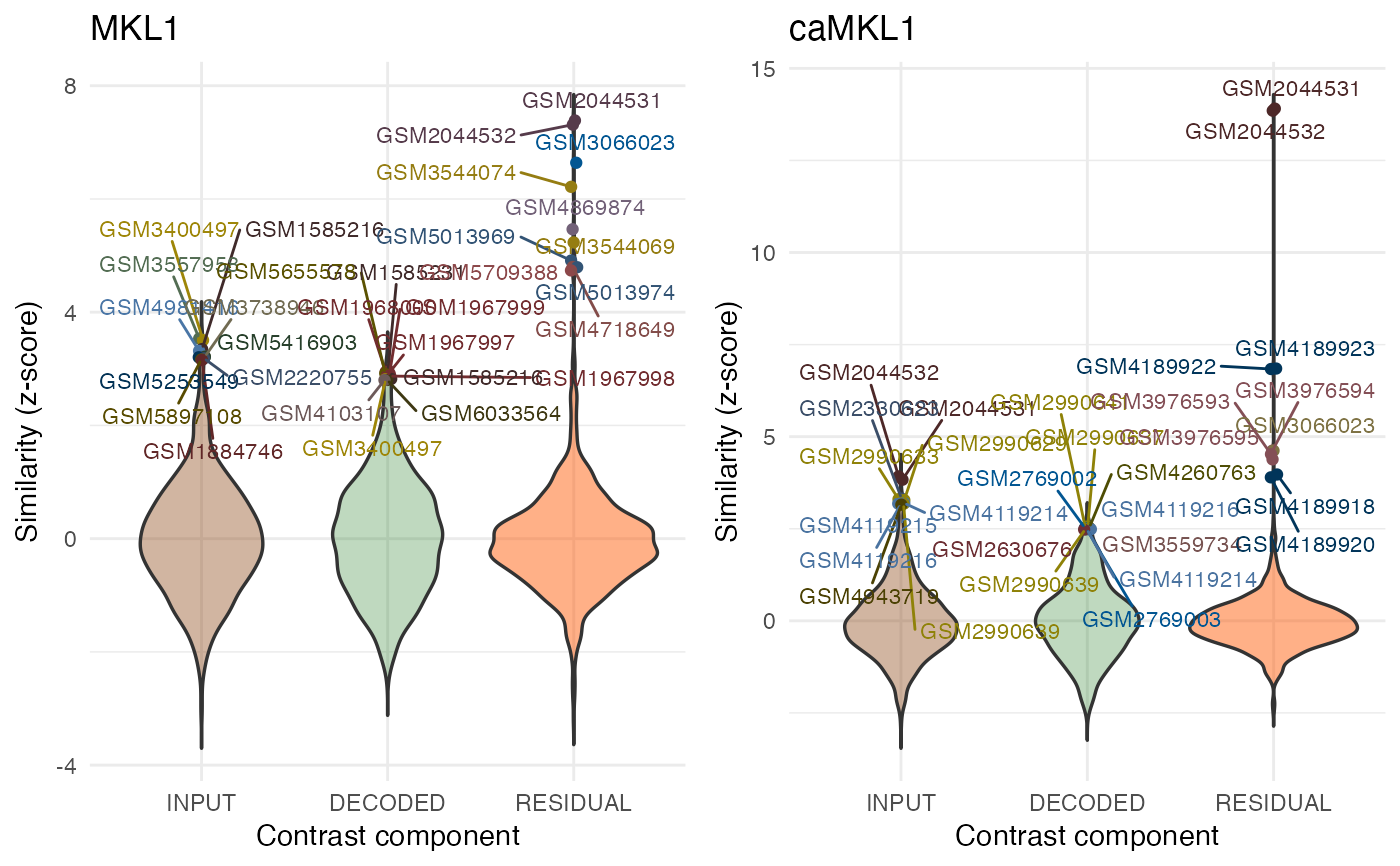
Select the type of visualization for the query results
"violin", "manh"or"none"to suppress the plotting.- detailTopn
specifies the number of top hits for which metadata will be returned in the TopHits slot of the results.
- verbose
Logical scalar indicating whether to print messages along the way.
- BPPARAM
BiocParallelParam object specifying how parallelization is to be performed using e.g.
MulticoreParam) orSnowParam).- chunk_size
Column dimension for the grid used to read blocks from the HDF5 Matrix. Sizes between 250 and 1000 are recommended. Smaller sizes reduce memory usage.
- mode
When in "ANALYSIS" mode (default) the complete contrast DB is queried. "DEMO" mode employs a small "toy" database for the queries. "DEMO" should only be used for testing/demonstration purposes and never for actual analysis purposes.
Value
A list with three elements called "pearson.rhos", "zscores" and
"TopHits", containing raw and z-scored Pearson's rho correlation
coefficients between the query contrast(s) and the contrasts in the
database, as well as detailed metadata for the detailTopn best
hits.
Examples
MKL1_human <- readRDS(system.file("extdata", "GSE215150_MKL1_Human.rds",
package = "orthos"))
# Decompose contrasts:
dec_MKL1_human <- decomposeVar(M = MKL1_human, treatm = c(2, 3), cntr = c(1, 1),
organism = "Human", verbose = FALSE)
#> see ?orthosData and browseVignettes('orthosData') for documentation
#> loading from cache
#> see ?orthosData and browseVignettes('orthosData') for documentation
#> loading from cache
#> see ?orthosData and browseVignettes('orthosData') for documentation
#> loading from cache
# Perform query against contrast DB with the decomposed fractions.
# !!!Note!!! mode="DEMO" for demonstration purposes only.
params <- BiocParallel::MulticoreParam(workers = 2)
query.res.human <- queryWithContrasts(dec_MKL1_human, organism = "Human",
BPPARAM = params, verbose = FALSE,
mode = "DEMO")
#> demo_decomposed_contrasts_human_rds already present in cache at: /Users/runner/Library/Caches/org.R-project.R/R/ExperimentHub/human_v212_NDF_c100_DEMOse.rds
#> demo_decomposed_contrasts_human_hdf5 already present in cache at: /Users/runner/Library/Caches/org.R-project.R/R/ExperimentHub/human_v212_NDF_c100_DEMOassays.h5