Filter out features in MaxQuant data
filterMaxQuant.RdExclude features with 'Score' below minScore, 'Peptides' below
minPeptides, or identified as either 'Reverse',
'Potential.contaminant' or 'Only.identified.by.site' by MaxQuant.
filterMaxQuant(sce, minScore, minPeptides, plotUpset = TRUE, exclFile = NULL)Arguments
- sce
A
SummarizedExperimentobject (or a derivative).- minScore
Numeric scalar, the minimum allowed value in the 'Score' column in order to retain the feature.
- minPeptides
Numeric scalar, the minimum allowed value in the 'Peptides' column in order to retain the feature.
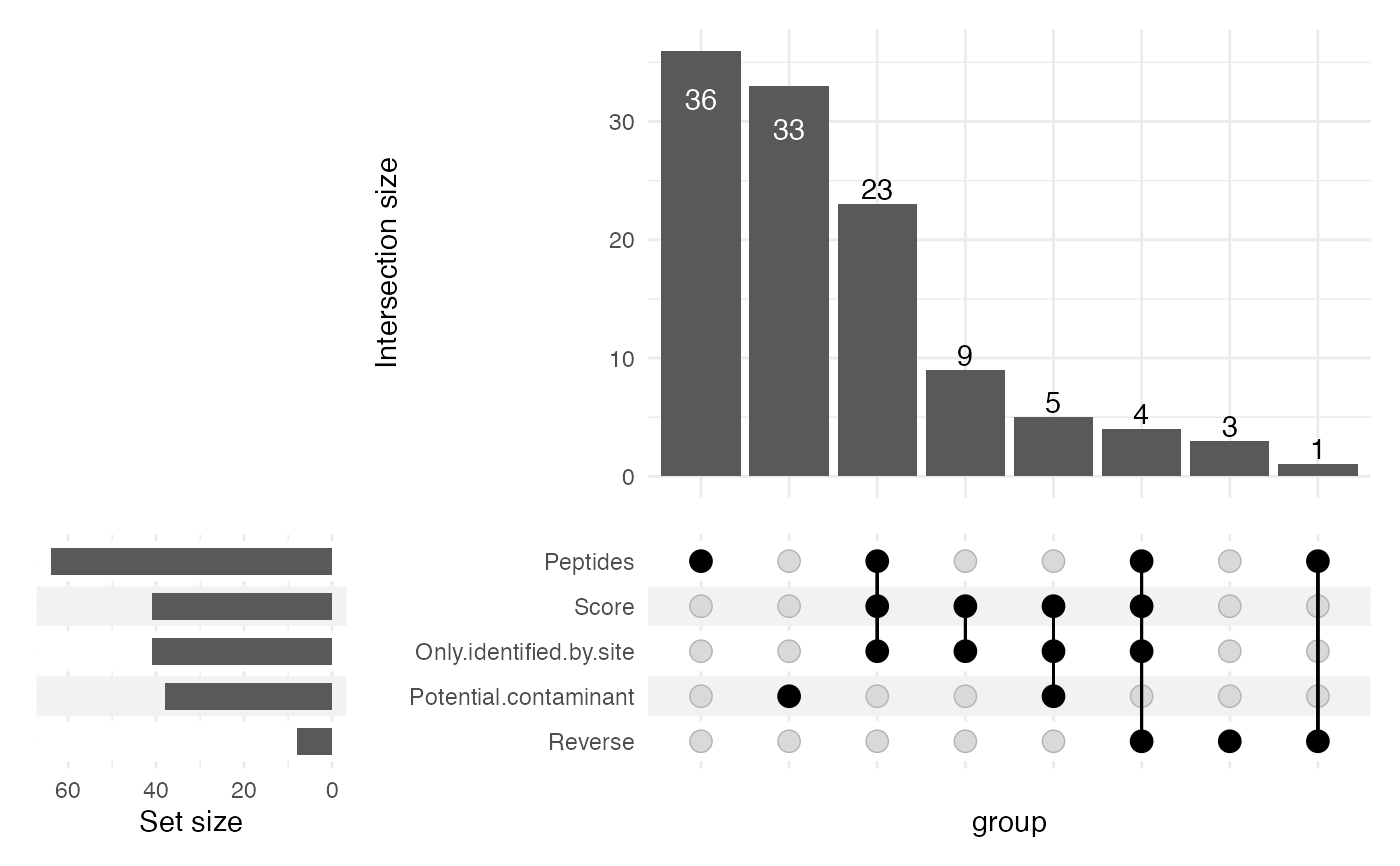
- plotUpset
Logical scalar, whether to generate an UpSet plot detailing the reasons for features being filtered out. Only generated if any feature is in fact filtered out.
- exclFile
Character scalar, the path to a text file where the features that are filtered out are written. If
NULL(default), excluded features are not recorded.
Value
A filtered object of the same type as sce.
Examples
sce <- importExperiment(inFile = system.file("extdata", "mq_example",
"1356_proteinGroups.txt",
package = "einprot"),
iColPattern = "^LFQ.intensity.")$sce
dim(sce)
#> [1] 463 9
sce <- filterMaxQuant(sce = sce, minScore = 2, minPeptides = 2,
plotUpset = TRUE)
 dim(sce)
#> [1] 349 9
dim(sce)
#> [1] 349 9