Filter out features in FragPipe data
filterFragPipe.RdExclude features with 'Combined.Total.Peptides' below minPeptides,
or identified as either 'Reverse' (Protein name matching
revPattern) or 'Potential.contaminant' (Protein name starting
with contam_) by FragPipe.
filterFragPipe(
sce,
minPeptides,
plotUpset = TRUE,
revPattern = "^rev_",
exclFile = NULL
)Arguments
- sce
A
SummarizedExperimentobject (or a derivative).- minPeptides
Numeric scalar, the minimum allowed value in the 'Combined.Total.Peptides' column in order to retain the feature.
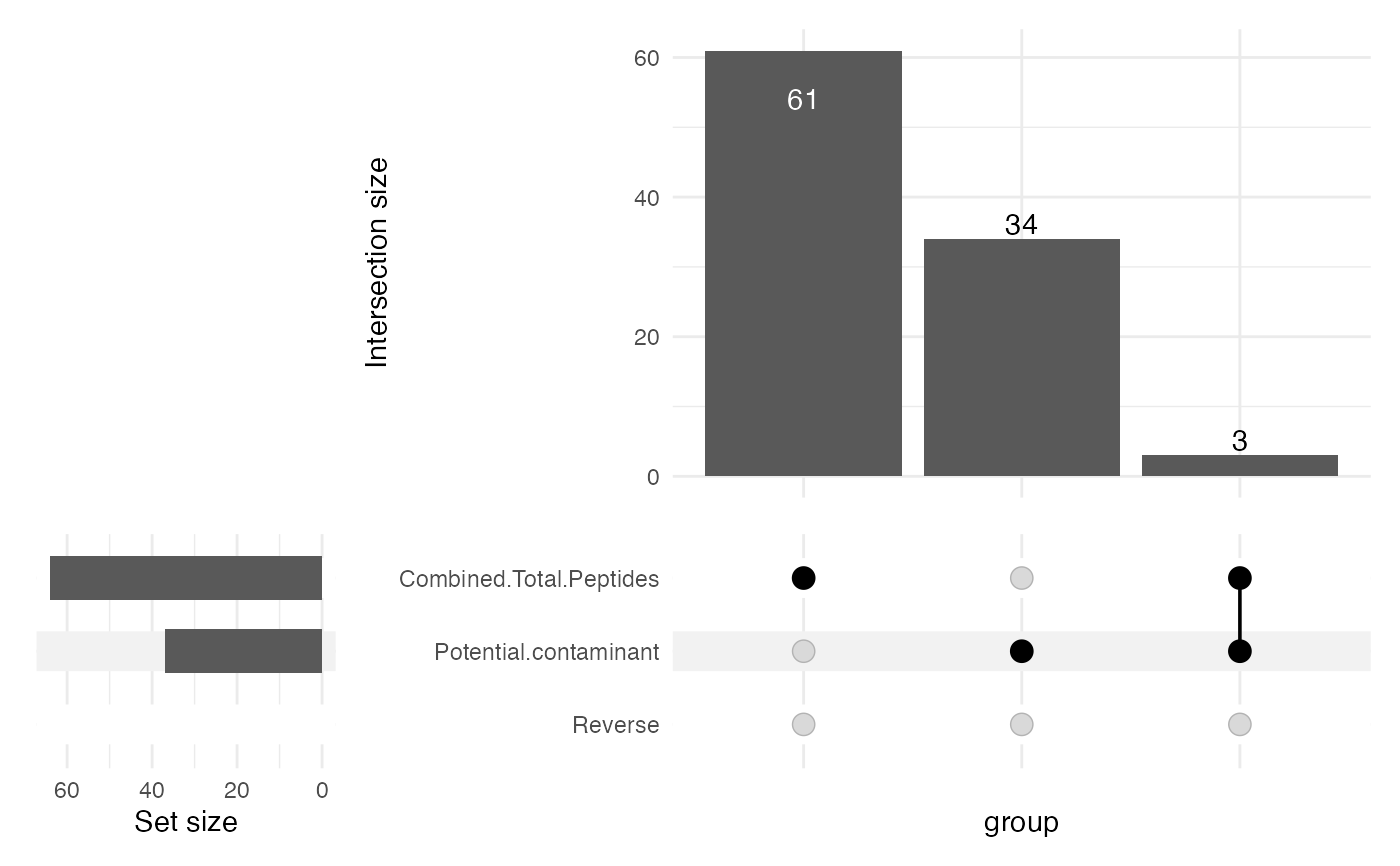
- plotUpset
Logical scalar, whether to generate an UpSet plot detailing the reasons for features being filtered out. Only generated if any feature is in fact filtered out.
- revPattern
Character scalar providing the pattern (a regular expression) used to identify decoys (reverse hits). The pattern is matched against the IDs in the FragPipe
Proteincolumn.- exclFile
Character scalar, the path to a text file where the features that are filtered out are written. If
NULL(default), excluded features are not recorded.
Value
A filtered object of the same type as sce.
Examples
sce <- importExperiment(inFile = system.file("extdata", "fp_example",
"combined_protein.tsv",
package = "einprot"),
iColPattern = ".MaxLFQ.Intensity$")$sce
dim(sce)
#> [1] 453 9
sce <- filterFragPipe(sce = sce, minPeptides = 2,
plotUpset = TRUE,
revPattern = "^rev_")
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the ComplexUpset package.
#> Please report the issue at
#> <https://github.com/krassowski/complex-upset/issues>.
 dim(sce)
#> [1] 355 9
dim(sce)
#> [1] 355 9